Learn about our research
Stem cell & Tissue engineering
Standardization of 3D stem cell cultures
We noticed that in stem cell cultures, the size of our 3D aggregates – or spheroids – was often dependent upon the culture conditions. We reasoned that in order to understand how stem cells react to their environment, we needed to more systematically decouple spheroid size and formation from differentiation. Micro-well technologies enable us to reproducibly control spheroid size and standardize differentiation.
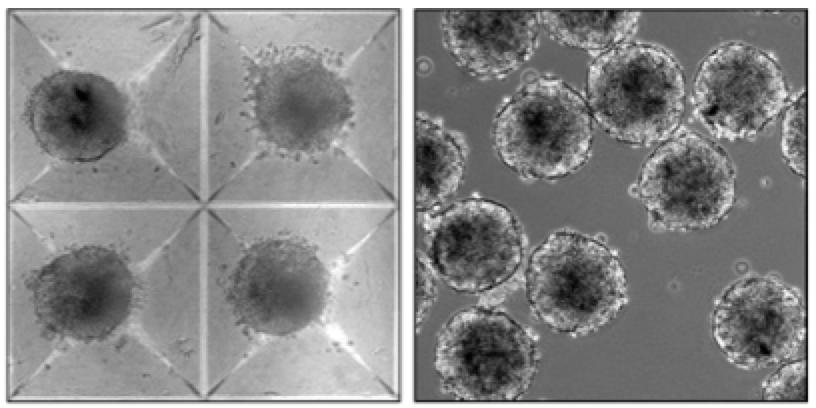
Influence of the stem cell environment
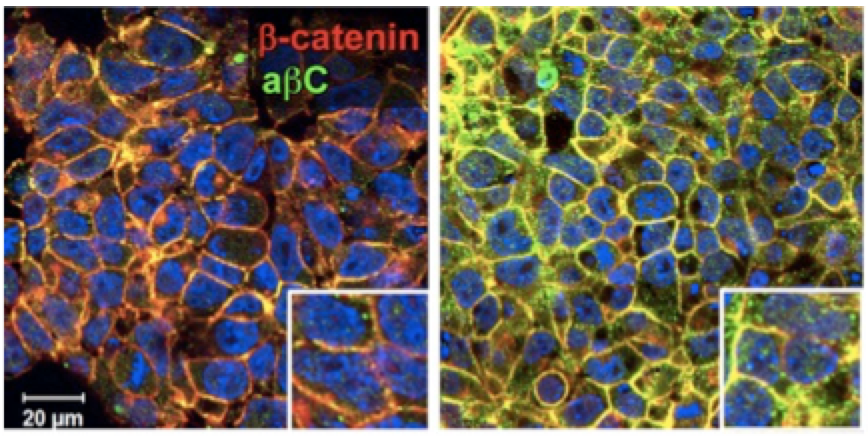
The sensitivity of stem cells to environmental perturbations prompted us to study the quantitative influences of spheroid culture conditions on differentiation. Suspension cultures that employ mixing of the media are routinely employed for large scale biomanufacturing applications. Strikingly, this relatively simple change to the culture conditions profoundly influences stem cell biology, including the interactions between cells, via the altered formation of cell-cell adhesions, downstream signaling, via Wnt/β-catenin, and ultimately the decision of cells to become cardiovascular.
Patterning and structure inside spheroids
Spheroids are not only influenced by their external environment, but also create and remodel an exquisitely complex cell-cell organization internally. When differentiated toward divergent lineages, the structure looks strikingly different. For example, when treated with BMP4, which directs cells toward mesodermal (i.e. blood, heart, etc.) lineages, cells exhibit a less dense structure, reminiscent of an epithelial-to-mesenchymal transition. Changes in cytoskeletal reorganization and ECM deposition that accompanies this process are also reflected through dynamic changes in the bulk mechanical properties and computational modeling suggested that these physical characteristics correlate strongly with the cell differentiation state.
Systems biology & hematology
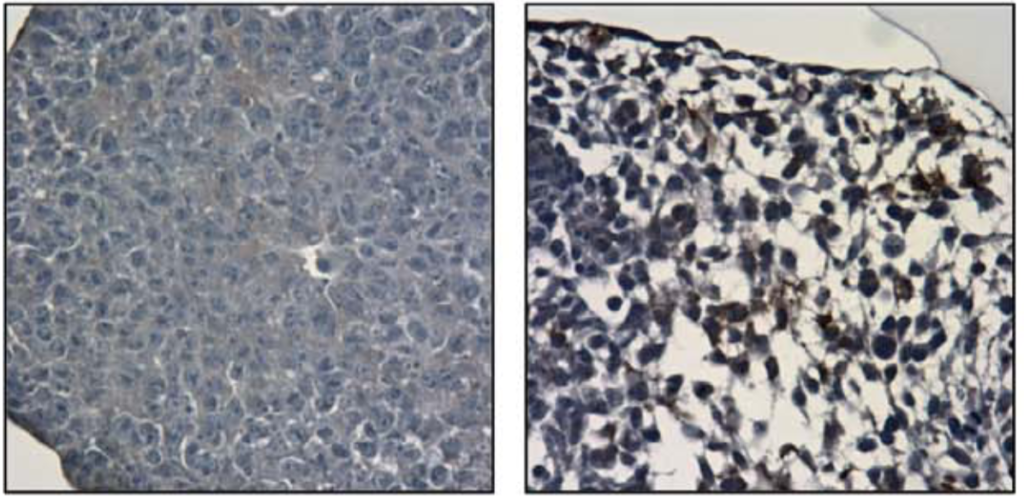
Epigenetic networks in T cell biology
Recent successes in immunotherapy have shown that T cells are an incredibly promising cell source for clinical applications. The robust generation of T cells from iPS cells would represent a major advance. Previous work had created an iPS-derived blood cell that was particularly compatible with the scale necessary for biomanufacturing cell therapies. These cells were restricted from becoming T cells; however, by altering epigenetic factors – specifically, EZH1 – we were able to “unlock” this T cell potential. This intriguing finding led us to investigate the networks of genes downstream of EZH1 by integrating RNA-, ATAC- and ChIP-seq data, which uncovered different targets and roles in blood stem cells (HSCs) and T cells.
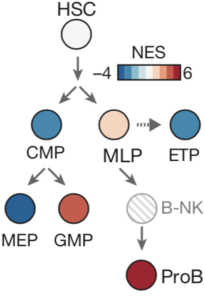
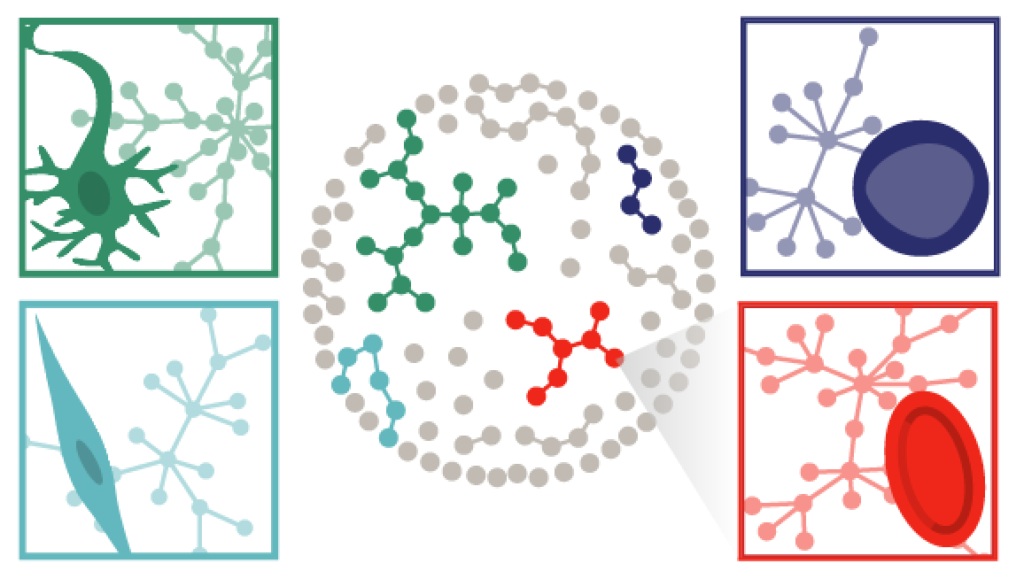
Connecting genes, pathways, and signaling
A fundamental question in stem cell and developmental biology is: what makes one cell type different from another? This question was addressed through CellNet, a pioneering computational algorithm. Building upon that foundation, we wanted to interrogate the differences at a more fine resolution between related cell types, such as those undergoing differentiation decisions. Using systems biology approaches, we created models of blood cells and the networks that mediate the transitions between stages. These models led us to identify a novel pathway, ErbB4, involved in the generation of red blood cells, and can be more broadly applied for engineering other cell types, including T cells.
Want to know more?
Melissa A. Kinney, Ph.D.
Department Of Biomedical Engineering
University Of Wisconsin-Madison
Engineering Centers Building
Room 2148
1550 Engineering Dr
Madison, WI 53706

